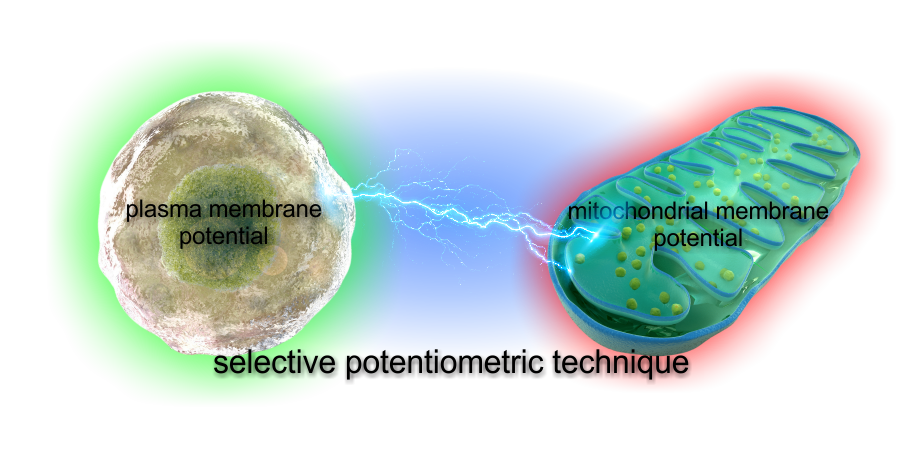
Absolute and Unbiased Mitochondrial Membrane Potential Assay
Commonly used mitochondrial membrane potential (ΔψM) assays often lead to data misinterpretation. This is because all of the used probes are influenced by multiple properties of cells other than ΔψM.
Our technology is based on biophysical modeling of ΔψM probe behavior to
back-calculate potentials from time courses of fluorescence
intensities. This enables absolute millivolts readout, and an
unbiased comparison of different samples, accounting for geometric,
binding and ΔψP effects.
The Theory
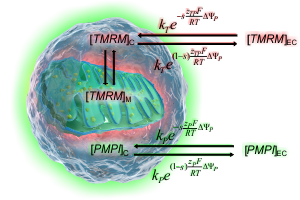
How absolute millivolts are calculated
See how effects of ΔψP, cell and mitochondrial geometry and fluorescence background accounted for to calculate mV values.
MoreIn Practice

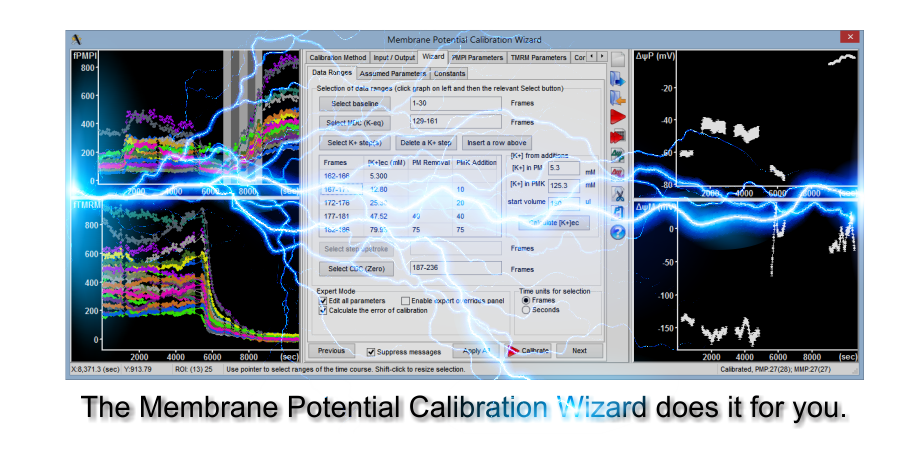
The Membrane Potential Calibration Wizard
This interactive dialog works hand in hand with the provided protocol to calibrate potentials without theoretical knowledge.
MoreTry it out
See the technology in publications
Mitochondrial Membrane Potential Assay Protocol

| The development of the unbiased, absolute mitochondrial membrane potential assay has been supported by: |

 |